DEPT
DEPT (Distortionless Enhancement by
Polazization Transfer) is a 13C detected,
multiplicity analysis experiment that differentiates CH, CH2 and CH3 carbons
from each other. Quaternary carbons are suppressed in the experiment. Varian
supplies a single DEPT pulse sequence (Dept). The editing is done in the pulse
sequence via a 1H pulse set to a 45, 90, or 135 degree rotation. The actual
setting is through a multiplier mult to a 90 degree pulse as
shown in the table below:
| Rotation (degree) |
mult |
Edited Results |
| 45 |
0.5 |
CH+CH2+CH3 (up) |
| 90 |
1.0 |
CH (up) |
| 135 |
1.5 |
CH+CH3 up (positive); CH2 down (negative) |
In the following,
as commonly found in literature, we will refer to the individual elements of
dept as dept45, dept90 and
dept135.
Varian
Dept Pulse Sequence
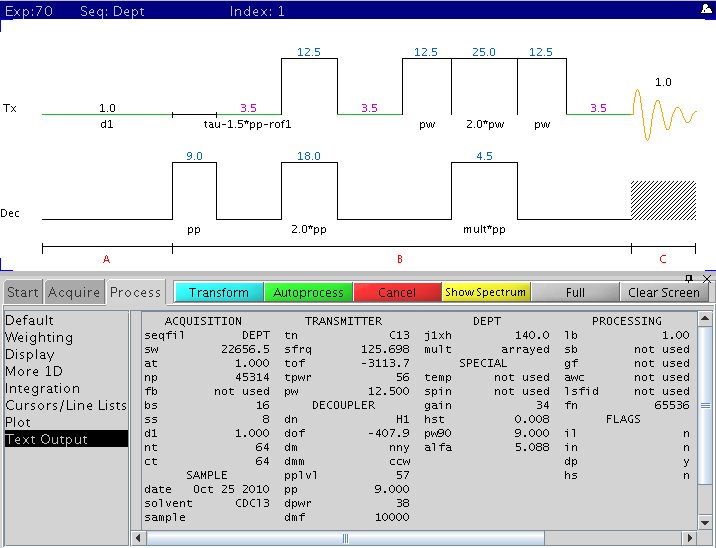
Note:
- DEPT requires accurate 90 degree 1H and to a lesser degree 13C 90 degree
pulse width to give a clean editing results.
- Varian's processing macro adept requires data with an
array of 4 experiments with mult=0.5, 1, 1, 1.5 with 1 run twice for even
sensitivity. The macro processes the raw data (wft adept)
to produce four individual spectra: All (CH+CH2+CH3), CH only, CH2 only,
and CH3 only.
- If you already have a 13C spectrum, it is often sufficient to run dept90
and dept135 separately in order to distinguish CH, CH2, and CH3 from each
other. In this case, judge from chemical shifts or other info to phase the
peaks up or down according to the table above. If desired, optionally run
dept45 to get the phase correct before phasing the dept90 and dept135
spectra.
- Optionally, you can run a customized 5-element array experiment
deptnoe (DEPT + 13C direct detection with NOE).
Procedure (on NMR500 with SWP probe)
Notes for all DEPT experiments:
- Turn ON temperature regulation (done by FTS on nmr500)
- Turn OFF spinning
- Cable connection remains the same as for 13C direct detection
- Tune both 1H channel and X-channel for 13C. This is
critical for clean editing.
- If some imperfection is to be tolerated in the final spectra, use the
default pulse widths. Otherwise, for best quality or if you observe
excessive poor editing results, calibrate 1H 90 degree pulse width with a
precision of 0.5 usec at tpwr=57. The default setting
assumes cdcl3 as solvent at 22C, but should also apply to solvents with
similar dielectric properties.
Option 1 (recommended)
- During Data Collection:
- Type procarray to process all
elements. Auto-phasing and scaling is applied. Spectra are displayed in
stacked plot mode.
- Alternatively, type wft
ds(1) f full to process all and display the 1st spectrum
after the 1st bs cycle is done. Type aph
to phase it and use manual phasing if needed. Once the 1st spectrum is
phased up, the phases of other spectra are set properly. Type wft
dssh dscale to display all four in horizontal
view, or wft dssa dscale in stacked plot
or vertical view. To shrink the peak heights to proper scaling, typing
vs=vs*2 or vs=vs*0.5 etc. followed by
dssa dscale.
- To separate the spectra CH, CH2 and CH3, type
adept after wft. Try wft adept
dssa.
- After Data Collection:
- To look at raw (unedited) data from each dept
element, follow the processing procedure above.
- If total spectral separation of CH, CH2 and CH3
is desired, click the autoprocess button . The macro
applies an automatic phasing, add/subtract the raw data into All, CH, CH2
and CH3 spectra. Note some add/subtract artifacts may be visible.
Alternatively, for manual processing, type wft adept to do
editing. After this command, type ds(1) f full aph
(manually correct phasing if necessary), followed by dssa.
Note adept should always act on data processed after
wft command. Always proceed adept with
wft.
- To print the either edited or unedited spectra,
type pldept.
- Or: type autodept to
auto-process and print both edited and edited DEPT.
Helpful Processing and Printing Commands
for DEPT
- procarray (auto-process, scale, and display array)
- plarray (plot array)
- autodept (auto-process and print both unedited and
edited DEPT spectra)
- pldept (print either unedited or edited DEPT
spectra)
- deptproc (performs DEPT editing, applies auto-phasing
and scaling, and display stacked plot)
- adept (performing DEPT editing only. No
auto-phasing/scaling or display is done. Always call wft
before adept)
Option 2 (For individual dept45, dept90, or dept135
runs)
- Lock and shim with spin off. Keep lock level at ~80%.
- Type setexp('dept') to load customized parameters for
DEPT experiment.
- Change solvent to yours
- Tune 1st channel to 13C and 2nd channel to 1H
- Optional: Change pw at tpwr=57 to
calibrated value if available
- To run dept90, type mult=1
- Or: to run dept135, type
mult=1.5
- Or: to run dept45, type
mult=0.5
- Or: to run dept90 and dept135 in one experiment
in an array mode, type: mult=1.0, 1.5
- Set nt to a large number multiple of 16
(nt=16000). For long unattended experiment, set
nt according to time available.
- Type go to start data collection.
- Default gain=34. Reduce it by 2 at a time if receiver
overflow light blinks on VT display panel, or ADC overflow message displays
above command line.
- Stop the experiment anytime the normal way by typing aa
or click the Abort Acquisition button.
- Processing:
- For single element data, type
wft. Apply manual phasing for dept90 and dept135 according
to Table 1 (DO NOT use autophasing for these two). For dept45, type
aph for auto-phasing (with manual adjustment if
necessary).
- For arrayed data, type
procarray for auto-processing and display. Or, type
wft f full dssa dscale to display arrayed spectra. Type
ds(1) f full to select the first one to phase.
Default parameters
- ss=8 nt=64 bs=16
- d1=1 at=1 gain=34 lb=1
- tpwr=56 pw=12.5 (for 13C)
- pplvl=57 pp=9.0 (for 1H, in cdcl3)
- mult=0.5, 1, 1, 1.5
- 13C center tof: ~ 75ppm, sw ~ 180ppm covering -15ppm to 165ppm.
- 1H center dof: ~ 4.2ppm with 1H decoupling during 13C aquisition
Example
- Sample: Strychnine at ~ 25mg/mL (~ 100mM) in
cdcl3
- Data collected October 2010 (Option 1, ~9 mins with nt=64)
|
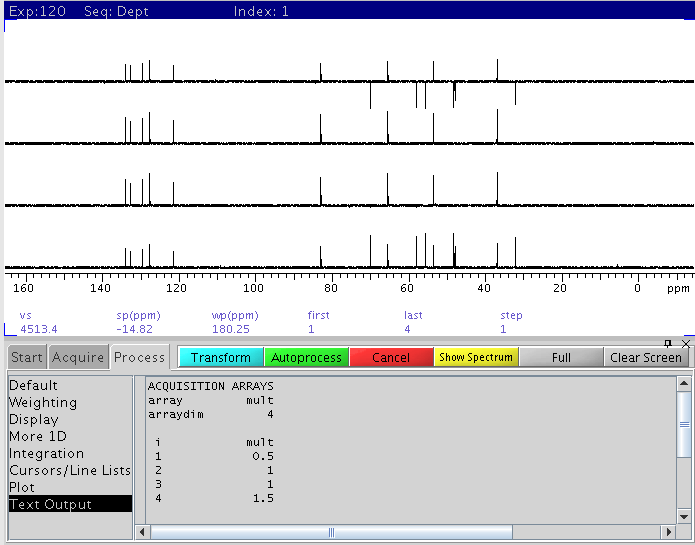
 |
| The 4 elements of Dept with mult=0.5, 1, 1, 1.5
(from bottom to top) correspond to dept45, dept90, dept90, and dept135.
These raw dept elements are sufficient for multiplicity analysis.
Further processing with adept is optional. |
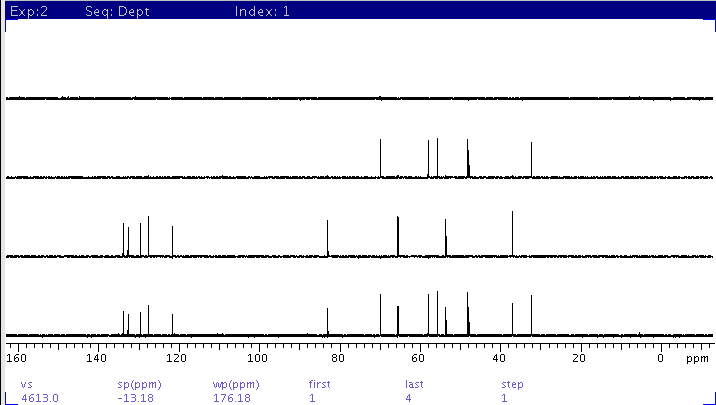
 |
| Spectra further processed with adept command. From
top to bottom:
1: CH3
2: CH2
3: CH
4: All (CH+CH2+CH3)
|
 |
|
|
H. Zhou
updated Oct 2010




