1H-1H
TOCSY
The following figure is the gradient based 2D TOCSY pulse sequence with the
main parameters displayed (NMR500).
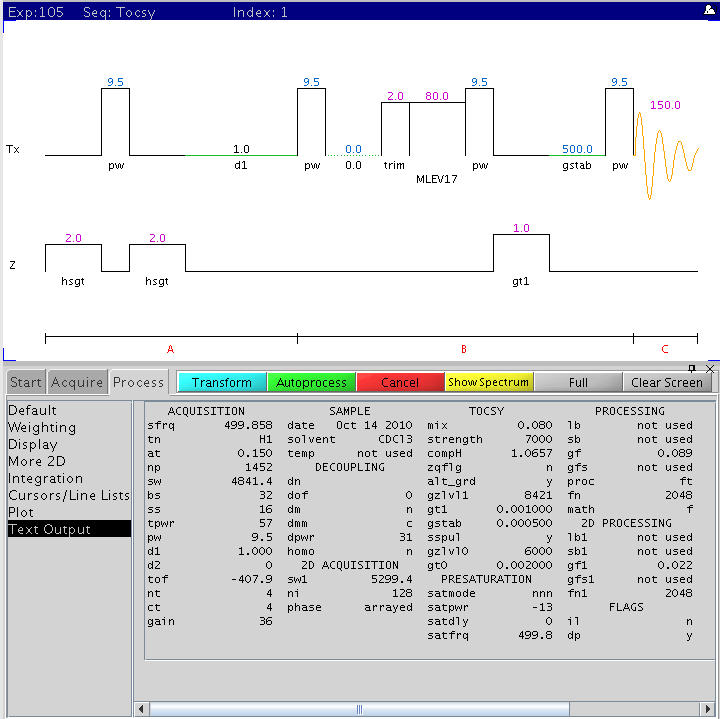
Procedure (on NMR500)
- Temperature must be regulated. Currently FTS is used to regulate
temperature. temp control in vnmrJ is turned off. Check VT operation procedure here if VT experiments are to be
done.
- Turn spin OFF.
- Gradients amplifier should be turned on (displays RUN).
Check the box sitting on the console. If front control button is on
Standby, flip it to ON.
- Type pfgon='nny' su to allow gradient pulses.
- Lock and shim your sample as usual. Keep lock at ~70-80% after
shimming.
- Find proper sw and tof to use
- Under exp1 (type jexp1 to go
there), collect a standard 1H spectrum (nt=1 is Ok for this, but 4 gives
cleaner spectrum)
- Put the box cursor to enclose the signal region
and to include ~10% flat baseline area at each end of the peak region. Type
movesw. This command sets a new spectral width
sw and spectrum center tof.
- Re-collect a 1D 1H spectrum with the new setting.
Type wft f full. Reference the 1H spectrum carefully.
- Load standard 2D TOCSY data file
- Join a different experiment (i.e.
jexp2 or create it with cexp(2) if it
doesn't exist).
- From exp2, type mf(1,2) to move
fid from exp1 to exp2. Type wft f full to display
spectrum. Check reference.
- Type setexp('tocsy')
su to load standard TOCSY parameters. Solvent, spectral
width, and center are copied over from 1D spectrum.
- Tune 1H channel. Accuracy of pre-calibrated pw depends
on probe tuning.
- Check/set proper 1H transmitter power and pulse width. If solvent is
CDCL3, tpwr=57 pw=9.5. For other solvents, calibrate pw
according to this
procedure.
- Use default parameters values (~20 mins experiment) except the
following:
- pw=XXX pw90=pw
(value calibrated with your solvent at tpwr=57)
- Optionally, for better quality data (may results
in longer data collection time):
- ss=32 (more dummy scans before
data collection)
- nt=8 (or 12, 16, 24, 28, ...
4*n) for more sensitivity and less artifacts
- ni=400 (better resolution along
indirect dimension)
- d1=2 (for less artifacts and
sensitivity, but doubles data collection time)
- Type time to check experiment time.
- Type go to collect data.
- Watch for ADC or receiver overflow:
- In arrayed experiments such as 2D's, a fixed
gain (not autogain) must be set. Watch the receiver overflow light and the
ADC overflow error message immediately after the experiment starts. If the
receiver overflows (red light blinks on VT display) or ADC overflow message
is seen above the command area anytime during the experiment except during
the dummy scan period, it is likely the data are ruined. Stop the
experiment and reduce gain by 2-4 at a time until the overflow disappears.
The default (gain=36) works most of the time unless there
is an extremely strong 1H signal in your sample. With a gain above 20, it
makes only small differences.
Default parameters
The default setting (modified from Varian's default values) in our parameter
set uses the following for a quick 2D COSY:
- ni=128 (128complex points along indirect dimension. For
better resolution, set ni bigger, ie.
ni=256)
- d1=1 (recycle delay between scans is 1 sec)
- ss=16 (16 dummy scans before actual data collection)
- nt=4 (4 scans)
- phase=1,2 (phase-sensitive mode in indirect
detection)
- gain=36 (reduce it by 2 at a time if receiver or ADC
overflows)
- sspul='y' (spin randomization before recycle delay with
homospoil gradients turned on)
- mix=0.080 (TOCSY spinlock mixing time is 80 msec)
Data Processing
You can use vnmrJ to process the data, but vnmrJ's graphics display for 2D
spectrum leaves much to be desired. A separate program is strongly recommended
for detailed processing and analysis.
One of the most widely used free processing/display software package for
multi-dimensional NMR data processing is nmrPipe/nmrDraw. This
program package has been installed on NMR500 and the data station. Follow this procedure to process data with nmrPipe/nmrDraw.
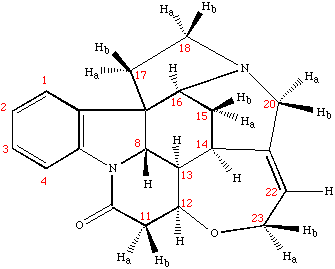
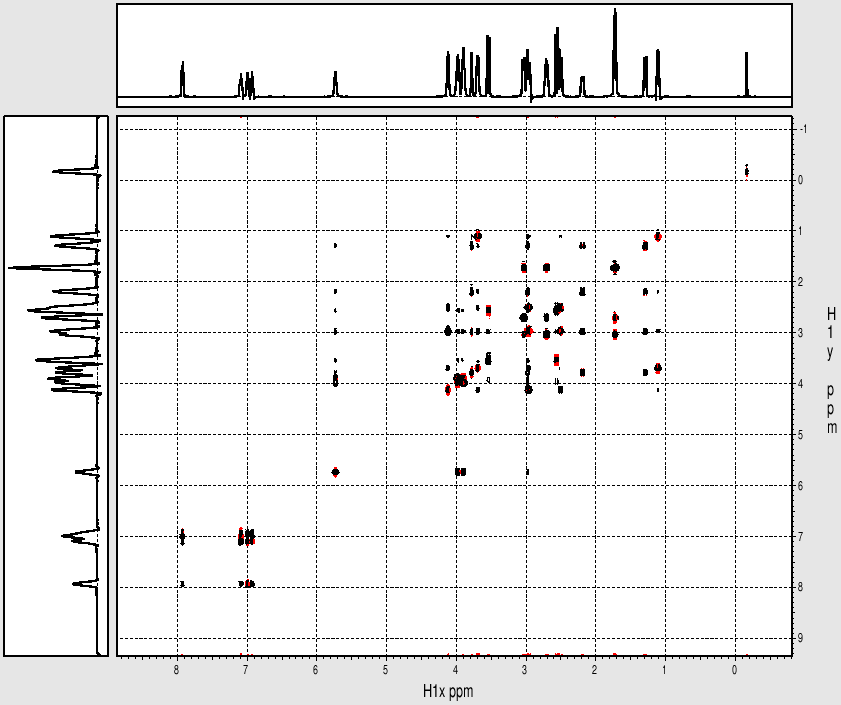
Example spectrum collected on NMR500
Sample: Strychnine at ~ 25mg/mL (~ 100mM) in cdcl3
- Data collected October 2010 (~21 mins with nt=4)
- TOCSY mixing time: 80 msec
- Processed and displayed with nmrPipe/nmrDraw
|
 |
H. Zhou
Updated Nov 2010



